You are here: Help » Searching » Genes » Detailed Query
Seaching (Genes)
Detailed Query
In order to find every bit of information for a few genes of interest and find out if the gene has been implicated to have an effect on any assay within the database we provide the Detailed Query search function. This scans the database and orders projects in terms of significance for each gene entered into the search field.
In order to use this tool, click the "Search" main menu option. Select the Detailed Query tab, and enter either a list of genes (RefSeq IDs, Flybase IDs or symbols) or Dharmacon siRNA IDs (seperated by white spage or commas). As this search query gathers information on ALL screens and projects within our database, there is a maximum of 5 search terms per query. The search only accepts either Dharmacon IDs OR geneIDs, it does not permit both types. Please note, if the gene symbol is not found, it might be that an alternative symbol is available in our database, so it is worth checking alternative gene names. If you have any problems, please feel free to contact the HTS lab.
Detailed Query Results
The results of each gene that is entered in the search are grouped together and displayed seperately (Figure 2).
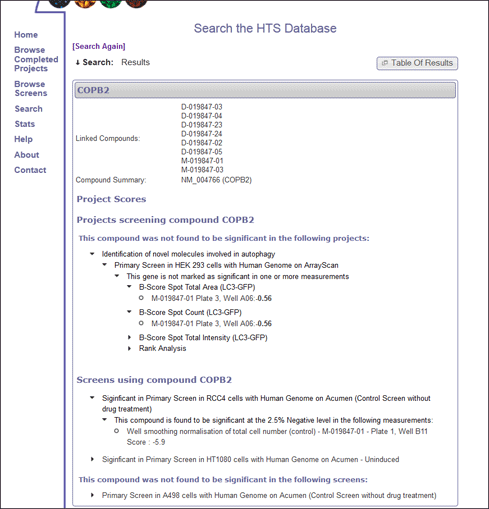
For each gene or compound, the scores of every dataset within every screen is identified and projects are ordered in terms of whether the gene or compound has a significant score in one or more screens within that project.
This allows the user to quickly identify projects where the gene or compound is marked as significant, and quickly discount, or focus on the relevant projects/screens.
Each project can be expanded to view the results of different screens and datasets within the project, where the well data scores can be displayed as required. A triangular icon before text indicates the statement can be expanded, and will either open a new page with more information, or expand to show extra information.
The compound search term appears in the top banner and links to the Library Compound View page.
The title of each project provides a link to an indepth view of the compound/gene scores within the project (Project Compound View), as well as providing more information about the project itself.
For more information on this view, please see workflow 4.
If you would like any help interpretting these results, please contact the HTS lab.